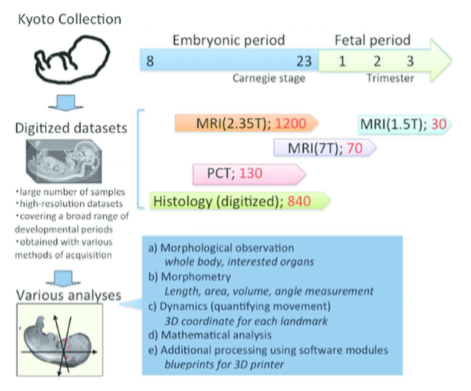
The Kyoto Collection samples were collected with well-structured, systematical methods and can be utilized for research purposes (Nishimura et al., 1968; Shiota, 1991). Most of these embryos and fetuses were obtained when pregnancy was terminated during the first trimester for socioeconomic reasons under the Maternity Protection Law of Japan. Some of the specimens (∼20%) were undamaged, well-preserved embryos. When the aborted embryos were brought to the laboratory, they were measured, examined, photographed, and their developmental stage was evaluated using the criteria provided by O’Rahilly and Müller (1987), the Carnegie stage (CS) system. Medical records were attached to most of the samples. Well-preserved human embryos and fetuses that were found to be externally normal were subjected to histological serial sectioning or digital data acquisition. Such disciplined methods for collecting the samples contribute to the selection of adequate samples for research.
The acquisition of digital datasets from the Kyoto Collection began in 1995 with histological serial sections observed using microscopy and captured with digital camera (Komori et al., 1995). Histological serial sections were then digitized using several methods. A high-resolution virtual slide scanner was used for preserving the glass slide materials, which are present, in part, on the web site (Kyoto Human Embryo Visualization Project, http://atlas.cac.med.kyoto-u.ac.jp). A lower-magnification film scanner with lower resolution (4800 dpi) was used for previewing the glass slides on computer screen and for 3D reconstructions. Histological serial sections of approximately 840 samples of both normal and abnormal embryos were obtained with the latter methods.
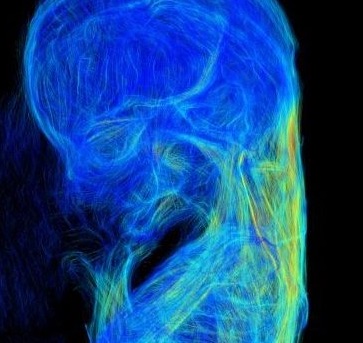
In 1999, Kyoto and Tsukuba Universities began a project to acquire 3D MR images of thousands of human embryos between CS13 and CS23 using a super-parallel MRI operated at 2.35 T, designated as MRI (2.35 T) in this review (Shiota et al., 2007; Matsuda et al., 2003, 2007). Namely, the MR images were acquired using about 100 samples per stage. To obtain 3D images of larger embryos in much higher resolution, a 7-T MR system, denoted MRI (7 T), (BioSpec 70/20 USR; Bruker Biospin MRI GmbH; Ettlingen, Germany) with a 35-mm-diameter 1H quadrature transmit-receive volume coil (T9988; Bruker Biospin MRI GmbH) was applied successively. Moreover, PCT, which is another principle image formation system, was also applied to obtain different data than high-resolution MRI. The details of the system and its features are described elsewhere (Yoneyama et al., 2006, 2004). For samples of fetal periods in the second and third trimesters, which have much larger size in the crown-rump length (CRL), MRI for clinical use was applied, denoted as MRI (1.5 T), (EXCELART Vantage, powered by Atlas; Toshiba Medical Systems, Otawara, Japan) with knee or head coils. Currently, approximately 70 samples (CRL 25–150 mm) of 3D images acquired by MRI (7 T), 30 samples of those in the second and third trimesters acquired by MRI (1.5 T), and 130 samples between CS14 and CS22 acquired by PCT were available for 3D analysis.
High-resolution 3D datasets with an adequately large number of samples, covering a broad range of developmental periods with various methods of acquisition, are key features for research. In particular, an adequate sample size is required for quantitative analysis using statistical methods. Since 2008, we have reported the 3D analysis of various organs by using the database. Such reports have proved that the digital datasets from Kyoto Collections are useful for research purposes (Figure 1, Table 1)